| assortativity | Obtain the assortativity coefficient for the given graph. |
| scalar_assortativity | Obtain the scalar assortativity coefficient for the given graph. |
| corr_hist | Obtain the correlation histogram for the given graph. |
| combined_corr_hist | Obtain the single-vertex combined correlation histogram for the given graph. |
| avg_neighbour_corr | Obtain the average neighbour-neighbour correlation for the given graph. |
| avg_combined_corr | Obtain the single-vertex combined correlation histogram for the given graph. |
Obtain the assortativity coefficient for the given graph.
| Parameters : | g : Graph
deg : string or PropertyMap
|
|---|---|
| Returns : | assortativity coefficient : tuple of two floats
|
See also
Notes
The assortativity coefficient [newman-mixing-2003] tells in a concise fashion how vertices of different types are preferentially connected amongst themselves, and is defined by
where \(a_i=\sum_je_{ij}\) and \(b_j=\sum_ie_{ij}\), and \(e_{ij}\) is the fraction of edges from a vertex of type i to a vertex of type j.
The variance is obtained with the jackknife method.
If enabled during compilation, this algorithm runs in parallel.
References
| [newman-mixing-2003] | M. E. J. Newman, “Mixing patterns in networks”, Phys. Rev. E 67, 026126 (2003), DOI: 10.1103/PhysRevE.67.026126 |
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... k = np.random.randint(1,max+1)
... accept = random() < 1.0/k
... return k
...
>>> g = gt.random_graph(1000, lambda: sample_k(40), model="probabilistic",
... vertex_corr=lambda i,k: 1.0 / (1 + abs(i - k)), directed=False,
... n_iter=100)
>>> gt.assortativity(g, "out")
(0.13903518011375607, 0.005051876804786422)
Obtain the scalar assortativity coefficient for the given graph.
| Parameters : | g : Graph
deg : string or PropertyMap
|
|---|---|
| Returns : | scalar assortativity coefficient : tuple of two floats
|
See also
Notes
The scalar assortativity coefficient [newman-mixing-2003] tells in a concise fashion how vertices of different types are preferentially connected amongst themselves, and is defined by
where \(a_x=\sum_ye_{xy}\) and \(b_y=\sum_xe_{xy}\), and \(e_{xy}\) is the fraction of edges from a vertex of type x to a vertex of type y.
The variance is obtained with the jackknife method.
If enabled during compilation, this algorithm runs in parallel.
References
| [newman-mixing-2003] | M. E. J. Newman, “Mixing patterns in networks”, Phys. Rev. E 67, 026126 (2003), DOI: 10.1103/PhysRevE.67.026126 |
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... k = np.random.randint(1,max+1)
... accept = random() < 1.0/k
... return k
...
>>> g = gt.random_graph(1000, lambda: sample_k(40), model="probabilistic",
... vertex_corr=lambda i,k: abs(i-k),
... directed=False, n_iter=100)
>>> gt.scalar_assortativity(g, "out")
(-0.44070158356400696, 0.010592022444678632)
>>> g = gt.random_graph(1000, lambda: sample_k(40), model="probabilistic",
... vertex_corr=lambda i, k: 1.0 / (1 + abs(i - k)),
... directed=False, n_iter=100)
>>> gt.scalar_assortativity(g, "out")
(0.6007430887839058, 0.011569809783643956)
Obtain the correlation histogram for the given graph.
| Parameters : | g : Graph
deg_source : string or PropertyMap
deg_target : string or PropertyMap
bins : list of lists (optional, default: [[0, 1], [0, 1]])
weight : edge property map (optional, default: None)
float_count : bool (optional, default: True)
|
|---|---|
| Returns : | bin_counts : ndarray
source_bins : ndarray
target_bins : ndarray
|
See also
Notes
The correlation histogram counts, for every vertex with degree (or scalar property) ‘source_deg’, the number of out-neighbours with degree (or scalar property) ‘target_deg’.
If enabled during compilation, this algorithm runs in parallel.
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... k = np.random.randint(1,max+1)
... accept = random() < 1.0/k
... return k
...
>>> g = gt.random_graph(10000, lambda: sample_k(40), model="probabilistic",
... vertex_corr=lambda i, j: (sin(i / pi) * sin(j / pi) + 1) / 2,
... directed=False, n_iter=100)
>>> h = gt.corr_hist(g, "out", "out")
>>> clf()
>>> xlabel("Source out-degree")
<...>
>>> ylabel("Target out-degree")
<...>
>>> imshow(h[0].T, interpolation="nearest", origin="lower")
<...>
>>> colorbar()
<...>
>>> savefig("corr.pdf")
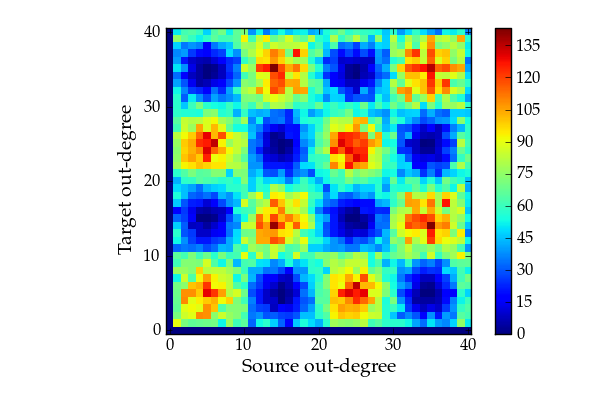
Out/out-degree correlation histogram.
Obtain the single-vertex combined correlation histogram for the given graph.
| Parameters : | g : Graph
deg1 : string or PropertyMap
deg2 : string or PropertyMap
bins : list of lists (optional, default: [[0, 1], [0, 1]])
float_count : bool (optional, default: True)
|
|---|---|
| Returns : | bin_counts : ndarray
first_bins : ndarray
second_bins : ndarray
|
See also
Notes
If enabled during compilation, this algorithm runs in parallel.
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... i = randint(1, max + 1)
... j = randint(1, max + 1)
... accept = random() < (sin(i / pi) * sin(j / pi) + 1) / 2
... return i,j
...
>>> g = gt.random_graph(10000, lambda: sample_k(40))
>>> h = gt.combined_corr_hist(g, "in", "out")
>>> clf()
>>> xlabel("In-degree")
<...>
>>> ylabel("Out-degree")
<...>
>>> imshow(h[0].T, interpolation="nearest", origin="lower")
<...>
>>> colorbar()
<...>
>>> savefig("combined_corr.pdf")
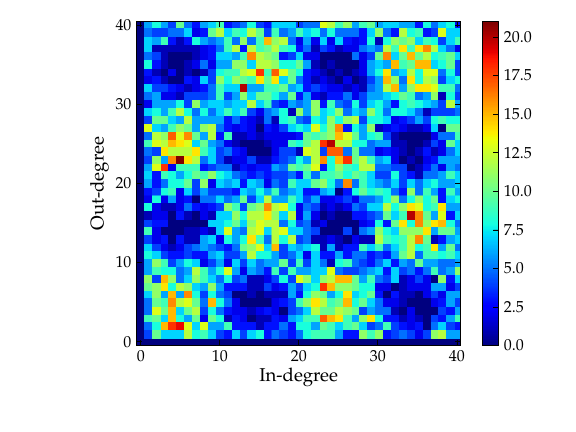
Combined in/out-degree correlation histogram.
Obtain the average neighbour-neighbour correlation for the given graph.
| Parameters : | g : Graph
deg_source : string or PropertyMap
deg_target : string or PropertyMap
bins : list (optional, default: [0, 1])
weight : edge property map (optional, default: None)
|
|---|---|
| Returns : | bin_avg : ndarray
bin_dev : ndarray
bins : ndarray
|
See also
Notes
The average correlation is the average, for every vertex with degree (or scalar property) ‘source_deg’, the of the ‘target_deg’ degree (or scalar property) of its neighbours.
If enabled during compilation, this algorithm runs in parallel.
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... k = randint(1,max+1)
... accept = random() < 1.0 / k
... return k
...
>>> g = gt.random_graph(10000, lambda: sample_k(40), model="probabilistic",
... vertex_corr=lambda i, j: (sin(i / pi) * sin(j / pi) + 1) / 2,
... directed=False, n_iter=100)
>>> h = gt.avg_neighbour_corr(g, "out", "out")
>>> clf()
>>> xlabel("Source out-degree")
<...>
>>> ylabel("Target out-degree")
<...>
>>> errorbar(h[2][:-1], h[0], yerr=h[1], fmt="o")
<...>
>>> savefig("avg_corr.pdf")
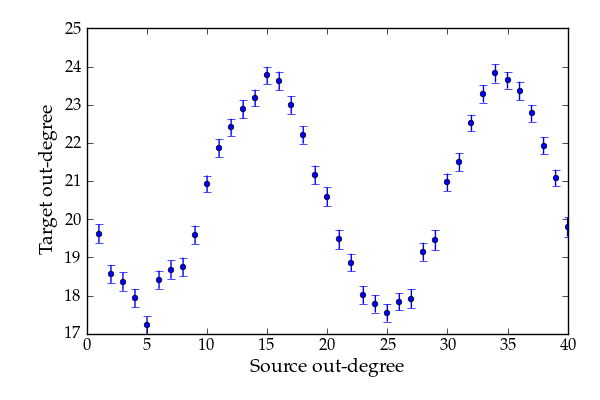
Average out/out degree correlation.
Obtain the single-vertex combined correlation histogram for the given graph.
| Parameters : | g : Graph
deg1 : string or PropertyMap
deg2 : string or PropertyMap
bins : list (optional, default: [0, 1])
|
|---|---|
| Returns : | bin_avg : ndarray
bin_dev : ndarray
bins : ndarray
|
See also
Notes
If enabled during compilation, this algorithm runs in parallel.
Examples
>>> def sample_k(max):
... accept = False
... while not accept:
... i = randint(1,max+1)
... j = randint(1,max+1)
... accept = random() < (sin(i/pi)*sin(j/pi)+1)/2
... return i,j
...
>>> g = gt.random_graph(10000, lambda: sample_k(40))
>>> h = gt.avg_combined_corr(g, "in", "out")
>>> clf()
>>> xlabel("In-degree")
<...>
>>> ylabel("Out-degree")
<...>
>>> errorbar(h[2][:-1], h[0], yerr=h[1], fmt="o")
<...>
>>> savefig("combined_avg_corr.pdf")
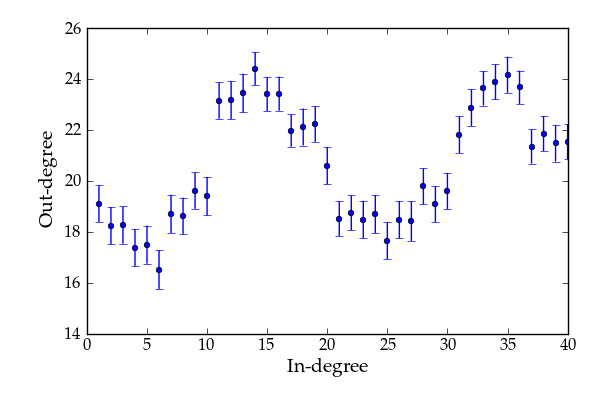
Average combined in/out-degree correlation.