This module contains algorithms for the computation of community structure on graphs.
| minimize_blockmodel_dl | Find the block partition of an unspecified size which minimizes the description length of the network, according to the stochastic blockmodel ensemble which best describes it. |
| BlockState | This class encapsulates the block state of a given graph. |
| mcmc_sweep | Performs a Monte Carlo Markov chain sweep on the network, to sample the block partition according to a probability \(\propto e^{-\beta \mathcal{S}_{t/c}}\), where \(\mathcal{S}_{t/c}\) is the blockmodel entropy. |
| collect_vertex_marginals | Collect the vertex marginal histogram, which counts the number of times a node was assigned to a given block. |
| collect_edge_marginals | Collect the edge marginal histogram, which counts the number of times the endpoints of each node have been assigned to a given block pair. |
| mf_entropy | Compute the “mean field” entropy given the vertex block membership marginals. |
| bethe_entropy | Compute the Bethe entropy given the edge block membership marginals. |
| model_entropy | Computes the amount of information necessary for the parameters of the traditional blockmodel ensemble, for B blocks, N vertices, E edges, and either a directed or undirected graph. |
| get_max_B | Return the maximum detectable number of blocks, obtained by minimizing: .. |
| get_akc | Return the minimum value of the average degree of the network, so that some block structure with \(B\) blocks can be detected, according to the minimum description length criterion. |
| min_dist | Return the minimum distance between all blocks, and the block pair which minimizes it. |
| condensation_graph | Obtain the condensation graph, where each vertex with the same ‘prop’ value is condensed in one vertex. |
| community_structure | Obtain the community structure for the given graph, using a Potts model approach. |
| modularity | Calculate Newman’s modularity. |
Find the block partition of an unspecified size which minimizes the description length of the network, according to the stochastic blockmodel ensemble which best describes it.
| Parameters : | g : Graph
deg_corr : bool (optional, default: True)
nsweeps : int (optional, default: 50)
adaptive_convergence : bool (optional, default: True)
anneal : float (optional, default: 1.)
greedy_cooling : bool (optional, default: True)
max_B : int (optional, default: None)
min_B : int (optional, default: 1)
mid_B : int (optional, default: None)
b_cache : dict with int keys and (float, PropertyMap) values (optional, default: None)
b_start : dict with int keys and (float, PropertyMap) values (optional, default: None)
clabel : PropertyMap (optional, default: None)
checkpoint : function (optional, default: None)
verbose : bool (optional, default: False)
|
|---|---|
| Returns : | b : PropertyMap
min_dl : float
b_cache : dict with int keys and (float, PropertyMap) values
|
Notes
This algorithm attempts to find a block partition of an unspecified size which minimizes the description length of the network,
where \(\mathcal{S}_{t/c}\) is the blockmodel entropy (as described in the docstring of mcmc_sweep() and BlockState.entropy()) and \(\mathcal{L}_{t/c}\) is the information necessary to describe the model (as described in the docstring of model_entropy() and BlockState.entropy()).
The algorithm works by minimizing the entropy \(\mathcal{S}_{t/c}\) for specific values of \(B\) via mcmc_sweep() (with \(\beta = 1\) and \(\beta\to\infty\)), and minimizing \(\Sigma_{t/c}\) via an one-dimensional Fibonacci search on \(B\). See [peixoto-parsimonious-2013] for more details.
This algorithm has a complexity of \(O(\tau E\ln B_{\text{max}})\), where \(E\) is the number of edges in the network, \(\tau\) is the mixing time of the MCMC, and \(B_{\text{max}}\) is the maximum number of blocks considered. If \(B_{\text{max}}\) is not supplied, it is computed as \(\sim\sqrt{E}\) via get_max_B(), in which case the complexity becomes \(O(\tau E\ln E)\).
References
| [holland-stochastic-1983] | Paul W. Holland, Kathryn Blackmond Laskey, Samuel Leinhardt, “Stochastic blockmodels: First steps”, Carnegie-Mellon University, Pittsburgh, PA 15213, U.S.A., DOI: 10.1016/0378-8733(83)90021-7 |
| [faust-blockmodels-1992] | Katherine Faust, and Stanley Wasserman. “Blockmodels: Interpretation and Evaluation.” Social Networks 14, no. 1-2 (1992): 5-61. DOI: 10.1016/0378-8733(92)90013-W |
| [karrer-stochastic-2011] | Brian Karrer, and M. E. J. Newman. “Stochastic Blockmodels and Community Structure in Networks.” Physical Review E 83, no. 1 (2011): 016107. DOI: 10.1103/PhysRevE.83.016107. |
| [peixoto-entropy-2012] | Tiago P. Peixoto “Entropy of Stochastic Blockmodel Ensembles.” Physical Review E 85, no. 5 (2012): 056122. DOI: 10.1103/PhysRevE.85.056122, arXiv: 1112.6028. |
| [peixoto-parsimonious-2013] | Tiago P. Peixoto, “Parsimonious module inference in large networks”, Phys. Rev. Lett. 110, 148701 (2013), DOI: 10.1103/PhysRevLett.110.148701, arXiv: 1212.4794. |
Examples
>>> g = gt.collection.data["polbooks"]
>>> b, mdl, b_cache = gt.minimize_blockmodel_dl(g)
>>> gt.graph_draw(g, pos=g.vp["pos"], vertex_fill_color=b, vertex_shape=b, output="polbooks_blocks_mdl.pdf")
<...>
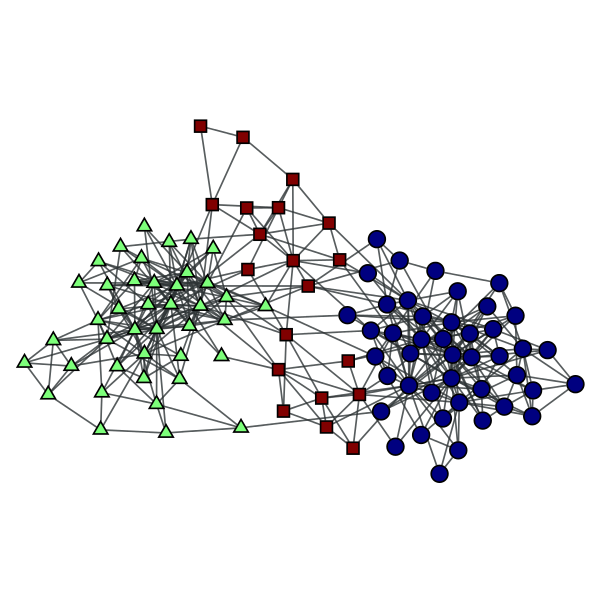
Block partition of a political books network, which minimizes the description lenght of the network according to the degree-corrected stochastic blockmodel.
This class encapsulates the block state of a given graph.
This must be instantiated and used by functions such as mcmc_sweep().
| Parameters : | g : Graph
eweight : PropertyMap (optional, default: None)
vweight : PropertyMap (optional, default: None)
b : PropertyMap (optional, default: None)
B : int (optional, default: None)
clabel : PropertyMap (optional, default: None)
deg_corr : bool (optional, default: True)
|
|---|
Add vertex v to block r.
Compute the “distance” between blocks r and s, i.e. the entropy difference after they are merged together.
Calculate the entropy per edge associated with the current block partition.
| Parameters : | complete : bool (optional, default: False)
random : bool (optional, default: False)
dl : bool (optional, default: False)
|
|---|
Notes
For the traditional blockmodel (deg_corr == False), the entropy is given by
for undirected and directed graphs, respectively, where \(e_{rs}\) is the number of edges from block \(r\) to \(s\) (or the number of half-edges for the undirected case when \(r=s\)), and \(n_r\) is the number of vertices in block \(r\) .
For the degree-corrected variant with “hard” degree constraints the equivalent expressions are
where \(e_r = \sum_se_{rs}\) is the number of half-edges incident on block \(r\), and \(e^+_r = \sum_se_{rs}\) and \(e^-_r = \sum_se_{sr}\) are the number of out- and in-edges adjacent to block \(r\), respectively.
If complete == False only the last term of the equations above will be returned. If random == True it will be assumed that \(B=1\) despite the actual \(e_{rs}\) matrix. If dl == True, the description length \(\mathcal{L}_t\) of the model will be returned as well, as described in model_entropy(). Note that for the degree-corrected version the description length is
where \(p_k\) is the fraction of nodes with degree \(p_k\), and we have instead \(k \to (k^-, k^+)\) for directed graphs.
Note that the value returned corresponds to the entropy per edge, i.e. \((\mathcal{S}_{t/c}\; [\,+\, \mathcal{L}_{t/c}])/ E\).
Returns the block graph.
Returns the property map which contains the block labels for each vertex.
Obtain the constraint label associated with each block.
Return the distance matrix between all blocks. The distance is defined as the entropy difference after two blocks are merged.
Returns the vertex property map of the block graph which contains the number \(e_r\) of half-edges incident on block \(r\). If the graph is directed, a pair of property maps is returned, with the number of out-edges \(e^+_r\) and in-edges \(e^-_r\), respectively.
Returns the edge property map of the block graph which contains the \(e_{rs}\) matrix entries.
Returns the block edge counts associated with the block matrix \(e_{rs}\). For directed graphs it is identical to \(e_{rs}\), but for undirected graphs it is identical except for the diagonal, which is \(e_{rr}/2\).
Returns the block matrix.
| Parameters : | reorder : bool (optional, default: False)
clabel : PropertyMap (optional, default: None)
niter : int (optional, default: 0)
ret_order : bool (optional, default: False)
|
|---|
Examples
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=5, deg_corr=True)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state)
>>> m = state.get_matrix(reorder=True)
>>> figure()
<...>
>>> matshow(m)
<...>
>>> savefig("bloc_mat.pdf")
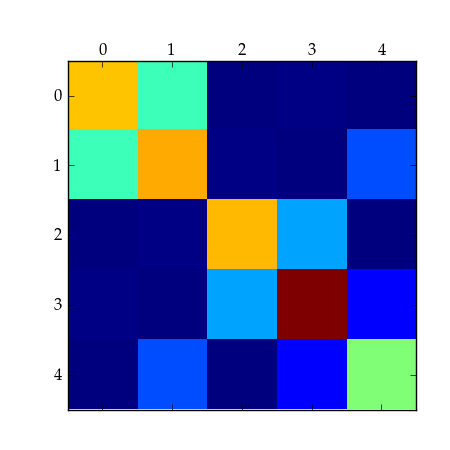
A 5x5 block matrix.
Returns the vertex property map of the block graph which contains the block sizes \(n_r\).
Merge blocks r and s into a single block.
Computes the modularity of the current block structure.
Move vertex v to block r, and return the entropy difference.
Remove vertex v from its current block.
Performs a Monte Carlo Markov chain sweep on the network, to sample the block partition according to a probability \(\propto e^{-\beta \mathcal{S}_{t/c}}\), where \(\mathcal{S}_{t/c}\) is the blockmodel entropy.
| Parameters : | state : BlockState
beta : float (optional, default: 1.0)
sequential : bool (optional, default: True)
verbose : bool (optional, default: False)
|
|---|---|
| Returns : | dS : float
nmoves : int
|
Notes
This algorithm performs a Monte Carlo Markov chain sweep on the network, where the block memberships are randomly moved, and either accepted or rejected, so that after sufficiently many sweeps the partitions are sampled with probability proportional to \(e^{-\beta\mathcal{S}_{t/c}}\), where \(\mathcal{S}_{t/c}\) is the blockmodel entropy, given by
for undirected and directed traditional blockmodels (deg_corr == False), respectively, where \(e_{rs}\) is the number of edges from block \(r\) to \(s\) (or the number of half-edges for the undirected case when \(r=s\)), and \(n_r\) is the number of vertices in block \(r\), and constant terms which are independent of the block partition were dropped (see BlockState.entropy() for the complete entropy). For the degree-corrected variant with “hard” degree constraints the equivalent expressions are
where \(e_r = \sum_se_{rs}\) is the number of half-edges incident on block \(r\), and \(e^+_r = \sum_se_{rs}\) and \(e^-_r = \sum_se_{sr}\) are the number of out- and in-edges adjacent to block \(r\), respectively.
The Monte Carlo algorithm employed attempts to improve the mixing time of the markov chain by proposing membership moves \(r\to s\) with probability \(p(r\to s|t) \propto e_{ts} + 1\), where \(t\) is the block label of a random neighbour of the vertex being moved. See [peixoto-parsimonious-2013] for more details.
This algorithm has a complexity of \(O(E)\), where \(E\) is the number of edges in the network.
References
| [holland-stochastic-1983] | Paul W. Holland, Kathryn Blackmond Laskey, Samuel Leinhardt, “Stochastic blockmodels: First steps”, Carnegie-Mellon University, Pittsburgh, PA 15213, U.S.A., DOI: 10.1016/0378-8733(83)90021-7 |
| [faust-blockmodels-1992] | Katherine Faust, and Stanley Wasserman. “Blockmodels: Interpretation and Evaluation.” Social Networks 14, no. 1-2 (1992): 5-61. DOI: 10.1016/0378-8733(92)90013-W |
| [karrer-stochastic-2011] | Brian Karrer, and M. E. J. Newman. “Stochastic Blockmodels and Community Structure in Networks.” Physical Review E 83, no. 1 (2011): 016107. DOI: 10.1103/PhysRevE.83.016107. |
| [peixoto-entropy-2012] | Tiago P. Peixoto “Entropy of Stochastic Blockmodel Ensembles.” Physical Review E 85, no. 5 (2012): 056122. DOI: 10.1103/PhysRevE.85.056122, arXiv: 1112.6028. |
| [peixoto-parsimonious-2013] | Tiago P. Peixoto, “Parsimonious module inference in large networks”, Phys. Rev. Lett. 110, 148701 (2013), DOI: 10.1103/PhysRevLett.110.148701, arXiv: 1212.4794. |
Examples
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=3, deg_corr=True)
>>> pv = None
>>> for i in range(1000): # remove part of the transient
... ds, nmoves = gt.mcmc_sweep(state)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state)
... pv = gt.collect_vertex_marginals(state, pv)
>>> gt.graph_draw(g, pos=g.vp["pos"], vertex_shape="pie", vertex_pie_fractions=pv, output="polbooks_blocks_soft.pdf")
<...>
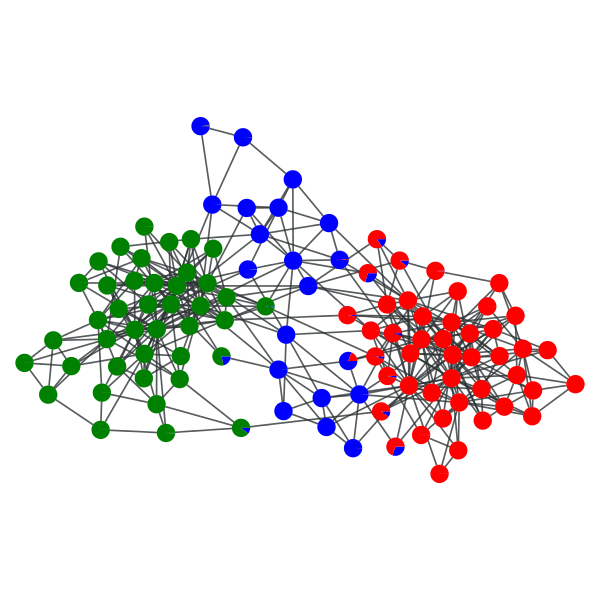
“Soft” block partition of a political books network with \(B=3\).
Collect the edge marginal histogram, which counts the number of times the endpoints of each node have been assigned to a given block pair.
This should be called multiple times, after repeated runs of the mcmc_sweep() function.
| Parameters : | state : BlockState
p : PropertyMap (optional, default: None)
|
|---|---|
| Returns : | p : PropertyMap (optional, default: None)
|
Examples
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=4, deg_corr=True)
>>> pe = None
>>> for i in range(1000): # remove part of the transient
... ds, nmoves = gt.mcmc_sweep(state)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state)
... pe = gt.collect_edge_marginals(state, pe)
>>> gt.bethe_entropy(state, pe)[0]
17.17007316834295
Collect the vertex marginal histogram, which counts the number of times a node was assigned to a given block.
This should be called multiple times, after repeated runs of the mcmc_sweep() function.
| Parameters : | state : BlockState
p : PropertyMap (optional, default: None)
|
|---|---|
| Returns : | p : PropertyMap (optional, default: None)
|
Examples
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=4, deg_corr=True)
>>> pv = None
>>> for i in range(1000): # remove part of the transient
... ds, nmoves = gt.mcmc_sweep(state)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state)
... pv = gt.collect_vertex_marginals(state, pv)
>>> gt.mf_entropy(state, pv)
20.001666525168361
>>> gt.graph_draw(g, pos=g.vp["pos"], vertex_shape="pie", vertex_pie_fractions=pv, output="polbooks_blocks_soft_B4.pdf")
<...>
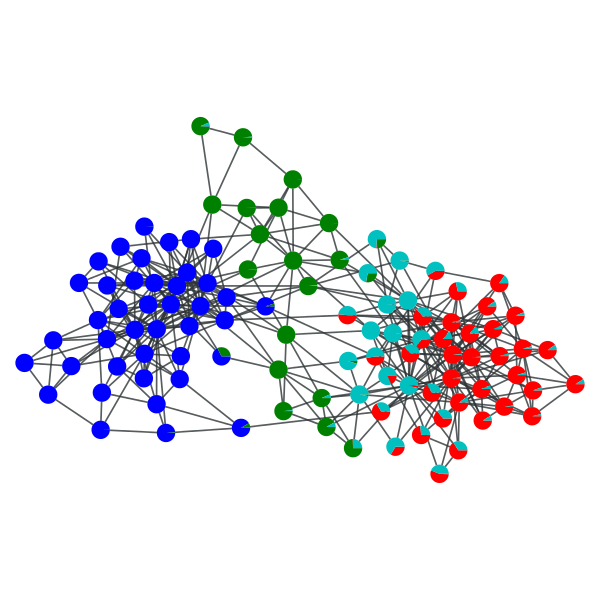
“Soft” block partition of a political books network with \(B=4\).
Compute the Bethe entropy given the edge block membership marginals.
| Parameters : | state : BlockState
p : PropertyMap
|
|---|---|
| Returns : | H : float
Hmf : float
pv : PropertyMap (optional, default: None)
|
Notes
The Bethe entropy is defined as,
where \(\pi_{(r,s)}^e\) is the marginal probability that the endpoints of the edge \(e\) belong to blocks \((r,s)\), and \(\pi_r^v\) is the marginal probability that vertex \(v\) belongs to block \(r\), and \(k_i\) is the degree of vertex \(v\) (or total degree for directed graphs).
References
| [mezard-information-2009] | Marc Mézard, Andrea Montanari, “Information, Physics, and Computation”, Oxford Univ Press, 2009. |
Compute the “mean field” entropy given the vertex block membership marginals.
| Parameters : | state : BlockState
p : PropertyMap
|
|---|---|
| Returns : | Hmf : float
|
Notes
The “mean field” entropy is defined as,
where \(\pi_r^v\) is the marginal probability that vertex \(v\) belongs to block \(r\).
References
| [mezard-information-2009] | Marc Mézard, Andrea Montanari, “Information, Physics, and Computation”, Oxford Univ Press, 2009. |
Computes the amount of information necessary for the parameters of the traditional blockmodel ensemble, for B blocks, N vertices, E edges, and either a directed or undirected graph.
A traditional blockmodel is defined as a set of \(N\) vertices which can belong to one of \(B\) blocks, and the matrix \(e_{rs}\) describes the number of edges from block \(r\) to \(s\) (or twice that number if \(r=s\) and the graph is undirected).
For an undirected graph, the number of distinct \(e_{rs}\) matrices is given by,
and for a directed graph,
where \(\left(\!{n \choose k}\!\right) = {n+k-1\choose k}\) is the number of \(k\) combinations with repetitions from a set of size \(n\).
The total information necessary to describe the model is then,
where \(N\ln B\) is the information necessary to describe the block partition.
References
| [peixoto-parsimonious-2013] | Tiago P. Peixoto, “Parsimonious module inference in large networks”, Phys. Rev. Lett. 110, 148701 (2013), DOI: 10.1103/PhysRevLett.110.148701, arXiv: 1212.4794. |
Return the maximum detectable number of blocks, obtained by minimizing:
where \(\mathcal{L}_t(B, N, E)\) is the information necessary to describe a traditional blockmodel with B blocks, N nodes and E edges (see model_entropy()).
References
| [peixoto-parsimonious-2013] | Tiago P. Peixoto, “Parsimonious module inference in large networks”, Phys. Rev. Lett. 110, 148701 (2013), DOI: 10.1103/PhysRevLett.110.148701, arXiv: 1212.4794. |
Examples
>>> gt.get_max_B(N=1e6, E=5e6)
1572
Return the minimum value of the average degree of the network, so that some block structure with \(B\) blocks can be detected, according to the minimum description length criterion.
This is obtained by solving
where \(\mathcal{L}_{t}\) is the necessary information to describe the blockmodel parameters (see model_entropy()), and \(\mathcal{I}_{t/c}\) characterizes the planted block structure, and is given by
where \(m_{rs} = e_{rs}/2E\) (or \(m_{rs} = e_{rs}/E\) for directed graphs) and \(w_r=n_r/N\). We note that \(\mathcal{I}_{t/c}\in[0, \ln B]\). In the case where \(E \gg B^2\), this simplifies to
for undirected and directed graphs, respectively. This limit is assumed if N == inf.
References
| [peixoto-parsimonious-2013] | Tiago P. Peixoto, “Parsimonious module inference in large networks”, Phys. Rev. Lett. 110, 148701 (2013), DOI: 10.1103/PhysRevLett.110.148701, arXiv: 1212.4794. |
Examples
>>> gt.get_akc(10, log(10) / 100, N=100)
2.4199998936261204
Return the minimum distance between all blocks, and the block pair which minimizes it.
The parameter state must be an instance of the BlockState class, and n is the number of block pairs to sample. If n == 0 all block pairs are sampled.
Examples
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=4, deg_corr=True)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state)
>>> gt.min_dist(state)
(795.7694502418633, 2, 3)
Obtain the condensation graph, where each vertex with the same ‘prop’ value is condensed in one vertex.
| Parameters : | g : Graph
prop : PropertyMap
vweight : PropertyMap (optional, default: None)
eweight : PropertyMap (optional, default: None)
avprops : list of PropertyMap (optional, default: None)
aeprops : list of PropertyMap (optional, default: None)
self_loops : bool (optional, default: False)
|
|---|---|
| Returns : | condensation_graph : Graph
prop : PropertyMap
vcount : PropertyMap
ecount : PropertyMap
va : list of PropertyMap
ea : list of PropertyMap
|
See also
Notes
Each vertex in the condensation graph represents one community in the original graph (vertices with the same ‘prop’ value), and the edges represent existent edges between vertices of the respective communities in the original graph.
Examples
Let’s first obtain the best block partition with B=5.
>>> g = gt.collection.data["polbooks"]
>>> state = gt.BlockState(g, B=5, deg_corr=True)
>>> for i in range(1000): # remove part of the transient
... ds, nmoves = gt.mcmc_sweep(state)
>>> for i in range(1000):
... ds, nmoves = gt.mcmc_sweep(state, beta=float("inf"))
>>> b = state.get_blocks()
>>> gt.graph_draw(g, pos=g.vp["pos"], vertex_fill_color=b, vertex_shape=b, output="polbooks_blocks_B5.pdf")
<...>
Now we get the condensation graph:
>>> bg, bb, vcount, ecount, avp, aep = gt.condensation_graph(g, b, avprops=[g.vp["pos"]], self_loops=True)
>>> gt.graph_draw(bg, pos=avp[0], vertex_fill_color=bb, vertex_shape=bb,
... vertex_size=gt.prop_to_size(vcount, mi=40, ma=100),
... edge_pen_width=gt.prop_to_size(ecount, mi=2, ma=10),
... output="polbooks_blocks_B5_cond.pdf")
<...>
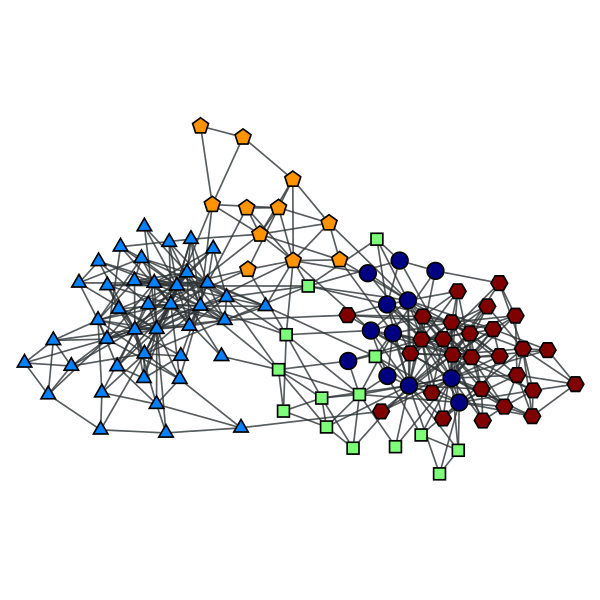
Block partition of a political books network with \(B=5\).
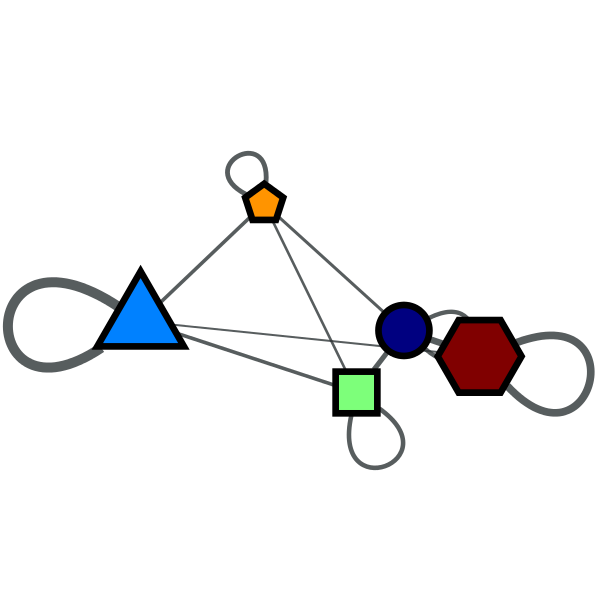
Condensation graph of the obtained block partition.
Obtain the community structure for the given graph, using a Potts model approach.
| Parameters : | g : Graph
n_iter : int
n_spins : int
gamma : float (optional, default: 1.0)
corr : string (optional, default: “erdos”)
spins : PropertyMap
weight : PropertyMap (optional, default: None)
t_range : tuple of floats (optional, default: (100.0, 0.01))
verbose : bool (optional, default: False)
history_file : string (optional, default: None)
|
|---|---|
| Returns : | spins : PropertyMap
|
See also
Notes
The method of community detection covered here is an implementation of what was proposed in [reichard-statistical-2006]. It consists of a simulated annealing algorithm which tries to minimize the following hamiltonian:
where \(p_{ij}\) is the probability of vertices i and j being connected, which reduces the problem of community detection to finding the ground states of a Potts spin-glass model. It can be shown that minimizing this hamiltonan, with \(\gamma=1\), is equivalent to maximizing Newman’s modularity ([newman-modularity-2006]). By increasing the parameter \(\gamma\), it’s possible also to find sub-communities.
It is possible to select three policies for choosing \(p_{ij}\) and thus choosing the null model: “erdos” selects a Erdos-Reyni random graph, “uncorrelated” selects an arbitrary random graph with no vertex-vertex correlations, and “correlated” selects a random graph with average correlation taken from the graph itself. Optionally a weight property can be given by the weight option.
The most important parameters for the algorithm are the initial and final temperatures (t_range), and total number of iterations (max_iter). It normally takes some trial and error to determine the best values for a specific graph. To help with this, the history option can be used, which saves to a chosen file the temperature and number of spins per iteration, which can be used to determined whether or not the algorithm converged to the optimal solution. Also, the verbose option prints the computation status on the terminal.
Note
If the spin property already exists before the computation starts, it’s not re-sampled at the beginning. This means that it’s possible to continue a previous run, if you saved the graph, by properly setting t_range value, and using the same spin property.
If enabled during compilation, this algorithm runs in parallel.
References
| [reichard-statistical-2006] | (1, 2) Joerg Reichardt and Stefan Bornholdt, “Statistical Mechanics of Community Detection”, Phys. Rev. E 74 016110 (2006), DOI: 10.1103/PhysRevE.74.016110, arXiv: cond-mat/0603718 |
| [newman-modularity-2006] | M. E. J. Newman, “Modularity and community structure in networks”, Proc. Natl. Acad. Sci. USA 103, 8577-8582 (2006), DOI: 10.1073/pnas.0601602103, arXiv: physics/0602124 |
Examples
This example uses the network community.xml.
>>> from pylab import *
>>> from numpy.random import seed
>>> seed(42)
>>> g = gt.load_graph("community.xml")
>>> pos = g.vertex_properties["pos"]
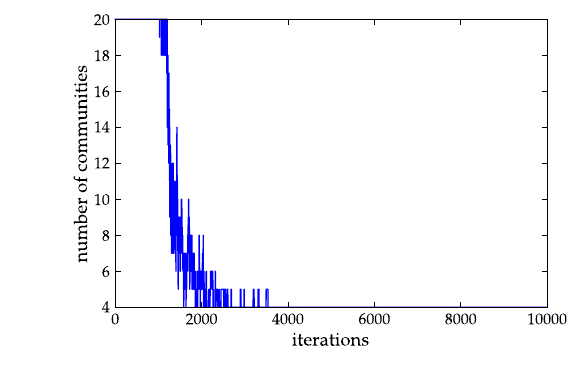
>>> spins = gt.community_structure(g, 10000, 20, t_range=(5, 0.1),
... history_file="community-history1")
>>> gt.graph_draw(g, pos=pos, vertex_fill_color=spins, output_size=(420, 420), output="comm1.pdf")
<...>
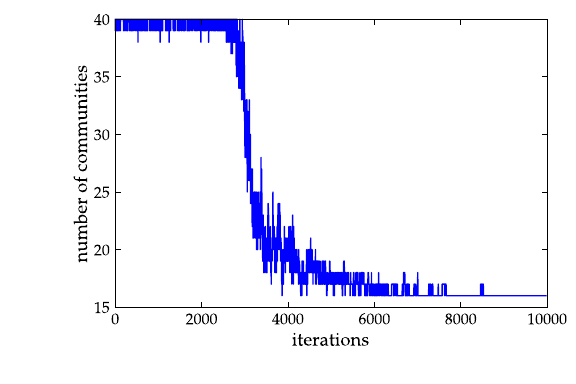
>>> spins = gt.community_structure(g, 10000, 40, t_range=(5, 0.1),
... gamma=2.5, history_file="community-history2")
>>> gt.graph_draw(g, pos=pos, vertex_fill_color=spins, output_size=(420, 420), output="comm2.pdf")
<...>
>>> figure(figsize=(6, 4))
<...>
>>> xlabel("iterations")
<...>
>>> ylabel("number of communities")
<...>
>>> a = loadtxt("community-history1").transpose()
>>> plot(a[0], a[2])
[...]
>>> savefig("comm1-hist.pdf")
>>> clf()
>>> xlabel("iterations")
<...>
>>> ylabel("number of communities")
<...>
>>> a = loadtxt("community-history2").transpose()
>>> plot(a[0], a[2])
[...]
>>> savefig("comm2-hist.pdf")
The community structure with \(\gamma=1\):
The community structure with \(\gamma=2.5\):

Calculate Newman’s modularity.
| Parameters : | g : Graph
prop : PropertyMap
weight : PropertyMap (optional, default: None)
|
|---|---|
| Returns : | modularity : float
|
See also
Notes
Given a specific graph partition specified by prop, Newman’s modularity [newman-modularity-2006] is defined by:
where \(e_{rs}\) is the fraction of edges which fall between vertices with spin s and r.
If enabled during compilation, this algorithm runs in parallel.
References
| [newman-modularity-2006] | M. E. J. Newman, “Modularity and community structure in networks”, Proc. Natl. Acad. Sci. USA 103, 8577-8582 (2006), DOI: 10.1073/pnas.0601602103, arXiv: physics/0602124 |
Examples
>>> from pylab import *
>>> from numpy.random import seed
>>> seed(42)
>>> g = gt.load_graph("community.xml")
>>> spins = gt.community_structure(g, 10000, 10)
>>> gt.modularity(g, spins)
0.535314188562404